On steganography
Isn’t that a cool word? It makes you think of dinosaurs doing dictation, or something. I first came across this word a few years ago when Bancroft and co-workers published a short letter in Nature, “Hiding messages in DNA microdots” (Nature, 399:533 (1999)). But steganography is actually a cryptography method in which you hide a signal amidst a great deal of noise or a great deal of other signal. For example, it is possible to hide messages within digital image files by having some subset of pixels actually encode a letter.
In any event, these authors recognized that steganography would be possible via DNA encoding, as well, and suggested “We have taken the microdot a step further and developed a DNA-based, doubly steganographic technique for sending secret messages. A DNA-encoded message is first camouflaged within the enormous complexity of human genomic DNA and then further concealed by confining this sample to a microdot.” I think there may even be a patent on this technique out there someplace.
But here’s the point: it’s only about ten years later, and this sounds so incredibly dated. It’s like poking Bulgarians with ricin-laced umbrellas, rather than properly exposing them to dioxin or polonium. And why is that? We all know the answer: NextGen sequencing. The enormous complexity of the human genome … is not really so enormous, anymore. Ten years of technology development and, poof, a reasonably clever idea is a joke. Sure, conceal your little oligonucleotide or plasmid or whatever in a bunch of human DNA. Bring it on! We’ll ferret out the unnatural or mutant sequence within, oh, about a day or so.
But let’s consider further. Is it really so silly to consider hiding messages in DNA? Well, actually, yes, given the enormous capabilities for information transfer available with electronics. Of course, there are concomitantly huge capabilities for codebreaking with electronics, and DNA does provide a convenient barrier to immediately tapping a digital format. And it is way more portable than a thumb drive. So, it is conceivable that there may be circumstances where DNA cryptography is a worthy pursuit.
And if so (a really big if) the question would become: how do we further hide the message, in this age of sequence-on-demand? In order to provide future generations (like, what, five years from now) with the ability to look back and chortle at my own foolishness, I suggest the following: alternative base-pairs.
I now must digress just a bit. DNA! I mean, wow! It is an impressive molecule. Not just because of what it does, but because of its incredible uniqueness. You, you there who have taken organic chemistry as an undergraduate, describe for me some other compound that can embed information in a similar way? Mrs. Robinson aside, I scoff at your plastics, which even in block copolymer form are mere random amalgams. I shake my head sadly at your host:guest complexes, which can barely interact specifically by halves, much less by millions or billions. And even the more viable biomolecule contenders, like peptides, are only exceptions that prove the rule. While Reza Ghadiri of Scripps is a stone-cold genius with his replicating leucine zippers, what they really prove to me is how very, very difficult it is to embed information in a replicating system, since the zippers form hypercycles that for the most part like to replicate towards an apotheosis, rather than towards the fixation of a random mutation.
DNA is unique. DNA is da bomb. Almost.
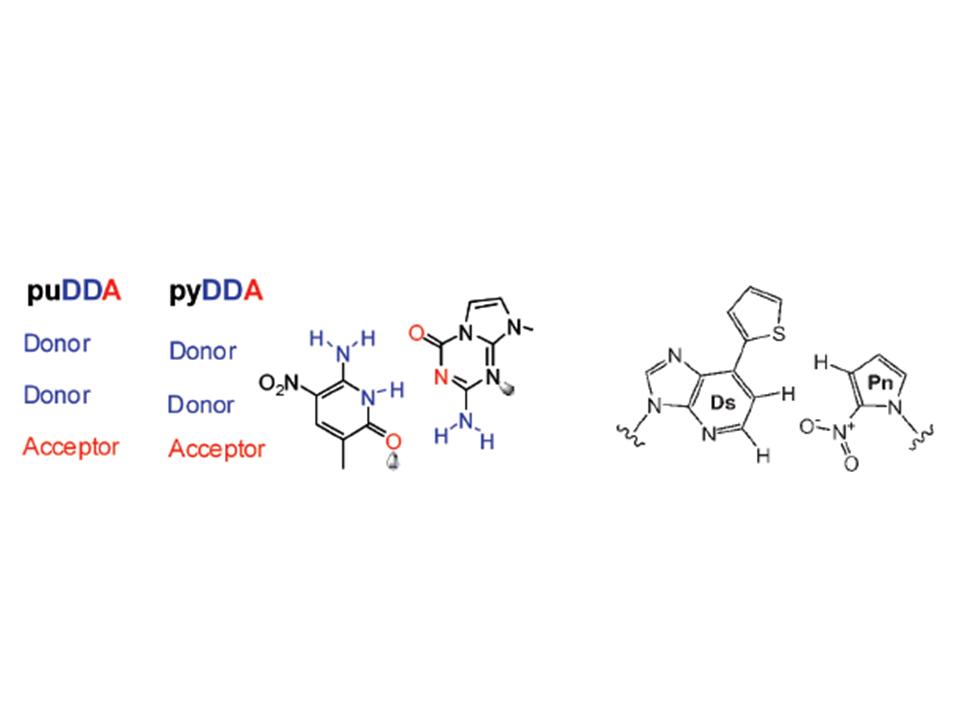
Proving that scientific brilliance can skip a generation, one of my scientific ‘fathers,’ Steve Benner (at the Foundation for Applied Molecular Evolution), and one of my scientific ’sons,’ Ichiro Hirao (at the Yokohama Institute, RIKEN), have designed, synthesized, and used PCR to incorporate non-natural base-pairs into DNA. Steve’s version (Yang et al. (2007), NAR 35:4238) is on the left, while Ichiro’s version (Kimoto et al. (2009), NAR 37:e14) is on the right, below. They have some of the same wonderful properties that DNA itself does: they’re isosteric with the natural pairs, they have bond complementarity and exclusivity, and they can fit into the same backbone that the other bases do. Now, you can sequence all the day long and you won’t find the strands that contain these weirdnesses, unless of course you happen to know in advance that you’re looking for them and have included the appropriate complementary nucleoside triphosphates in your sequencing reactions.
This leads to the question whether the probably limited number of available alternative base-pairs will be co-opted by particular intelligence services, in order to ensure complete secrecy of communications. In some future fantasy world will it be illegal to even describe some of these compounds? Um, no, for the reasons described above (electronics trumps biotechnology in this realm). And even so, we are still exploring whether bond complementarity is all that (go, go Eric Kool!) and whether other backbones are just as good as deoxyribose (a tip of the hat to Albert Eschenmoser). Still, never underestimate the creativity of scientists, and thus a truly facile replicating system that directly interfaces with electronics may yet arise. And if so, cryptography will be only one of its many, many applications.